Research Object Crate (RO-Crate) Update
2021-06-11

This presentation by Peter Sefton and Stian Soiland-Reyes was presented by Peter Sefton at the Open Repositories 2021 conference on 2021-06-10 (in Australia). RO-Crate has been presented at Open Repositories several times, including a workshop in 2019, so we won’t go through a very detailed introduction but we WILL start with with a quick introduction for those who have not seen it before.
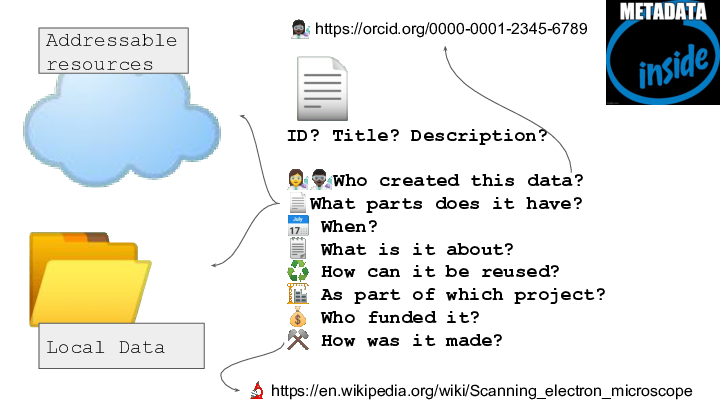
RO-Crate is method for describing a dataset as a digital object using a single linked-data metadata document which can have descriptions of files and resources that are local or remote, and can contain discipline-appropriate context for the data.
The dataset may contain any kind of data resource about anything, in any format as a file or URL
![3 📂
<p>|-- Folder1/
| |-- file1.this
| |-- file2.that
|-- Folder2/
| -- file1.this
| |-- file2.that
|-2021-04-08 07.58.17.jpg
{
"@id": "2021-04-08 07.58.17.jpg",
"@type": "File",
"contentSize": 3271409,
"dateModified": "2021-04-08T07:58:17+10:00",
"description": "",
"encodingFormat": [
{
"@id": "https://www.nationalarchives.gov.uk/PRONOM/x-fmt/391"
},
"image/jpeg"
],
"name": "Cute puppy"
},</p>
<p>](Slide3.png)
Each resource can have a machine readable description in JSON-LD format
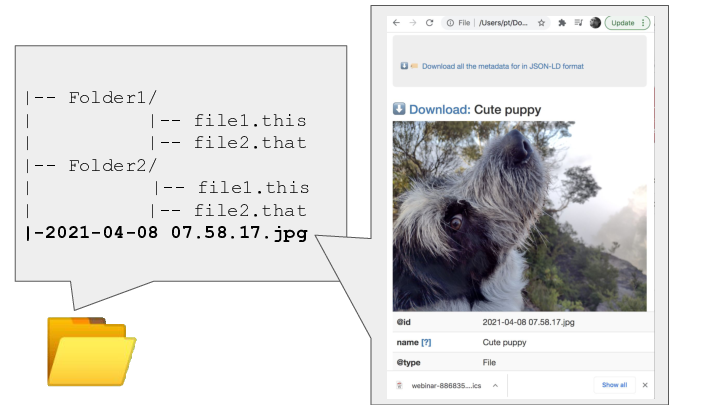
A human-readable description and preview can be in an HTML file that lives alongside the metadata
What does this mean for repositories? It means that a repository can show the contents of a digital object using either a standard display library, or a customised one.
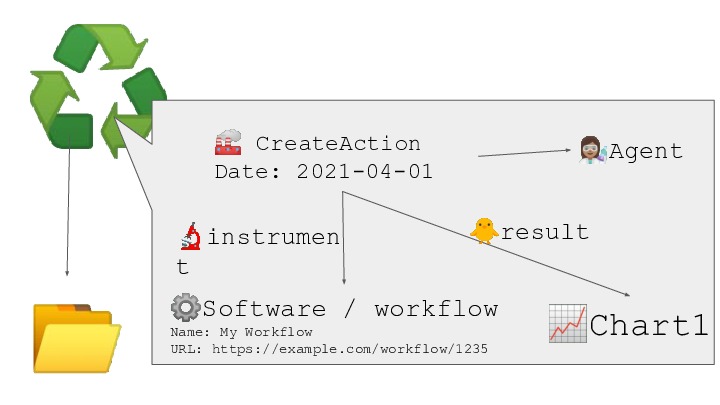
Provenance and workflow information can be included - to assist in data and research-process re-use.
What does this mean for repositories? Repositories will be able to launch software environments; if the digital object can be run in an emulator, or a notebook environment then there is potential to launch that.

RO-Crate Digital Objects may be packaged for distribution eg via Zip, Bagit and OCFL Objects.
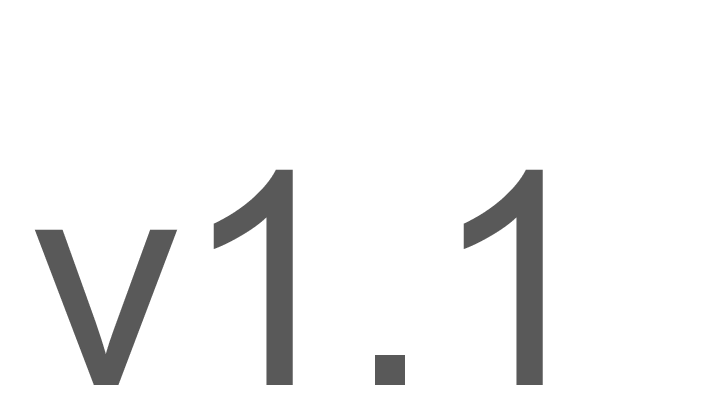
Since last Open Repositories we have reached V1.1. The main change are tidying up the file extension and making it clear that RO-Crates are not just packages of files - they are aggregations of local and remote objects, we’ll cover some other changes as well in the rest of the talk.
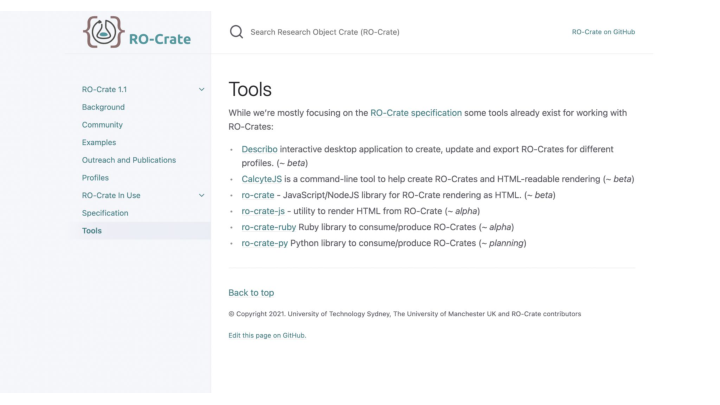
RO-Crate Tools keep coming.
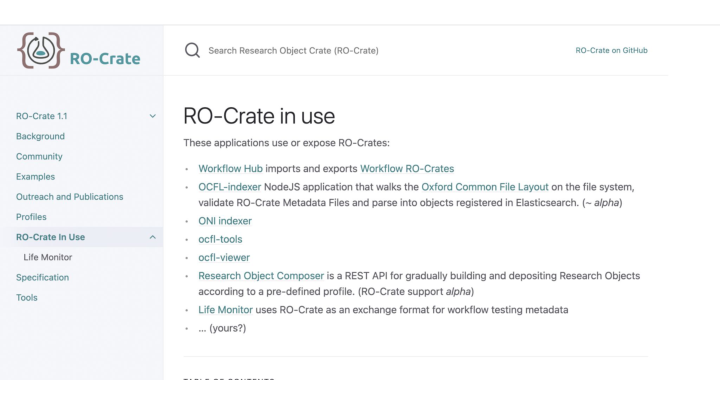
RO-Crate is being adopted in a number of projects

And RO-Crate is a foundation standard of the Arkisto platform - which was covered in the presentation before this one.
The RO-Crate team is now working on profiles - these will be guidance for humans and validation tools who want to use RO-Crate for specific purposes.
Image by Bryan Derksen - Original image Cup or faces paradox.jpg uploaded by Guam on 28 July 2005, SVG conversion by Bryan Derksen, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=1733355
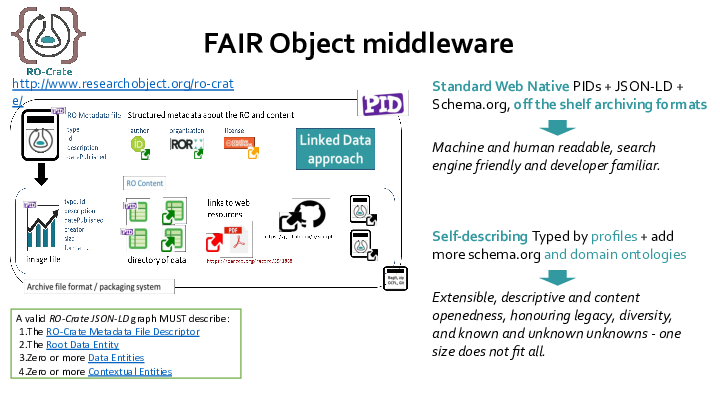
We are working on aligning RO-Crate with the work going on internationally on FAIR Digital Objects - coming from the standpoint of having a working FAIR-inspired way to create digital objects already.
From a forthcoming paper by Soiland-Reyes et al
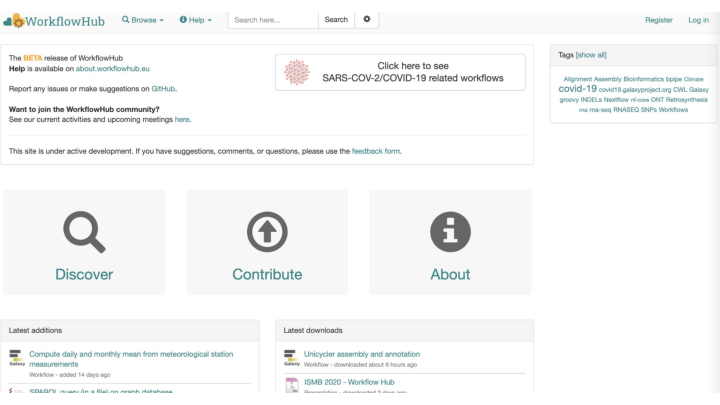
<WorkflowHub.eu> is a European cross-domain registry of computational workflows, supported by European Open Science Cloud projects, e.g. EOSC-Life, and research infrastructures including the pan-European bioinformatics network ELIXIR. As part of promoting workflows as reusable tools, WorkflowHub includes documentation and high-level rendering of the workflow structure independent of its native workflow definition format. The rationale is that a domain scientist can browse all relevant workflows for their domain, before narrowing down their workflow engine requirements. As such, the WorkflowHub is intended largely as a registry of workflows already deposited in repositories specific to particular workflow languages and domains, such as UseGalaxy.euand Nextflow nf-core .
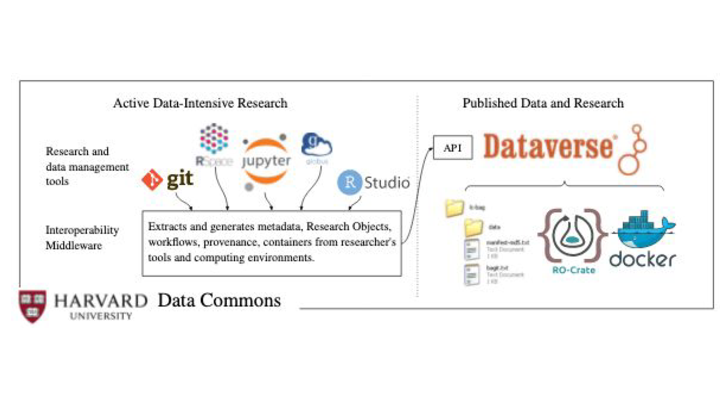
RO-Crate is featuring in discussions with Dataverse as a way of packing data.
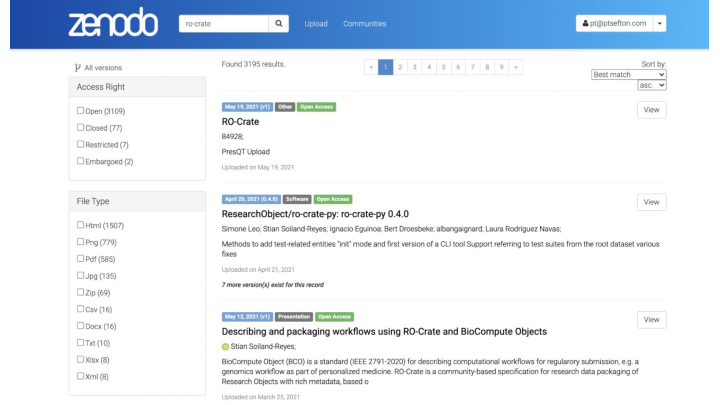
RO-Crate is going to be integrated with Zenodo as part of the CS3MESH4EOSC project, and by extension presumably the Invenio digital library framework. Of course RO-Crates can be deposited as they are can be wrapped as Zip files.

There are now enough things happening with RO-Crate that it is getting hard to keep track of it all - this slide is an incomplete view of what’s happening now.
RO-Crate is an open group - anyone can sign up - we have meetings twice a month that alternate between the European Morning and late evening / Australian late afternoon / Early morning.