Researchers now need to have end-to-end data management plans in place for all data, and all data must be archived appropriately to comply with the various codes and funding arrangements under which researchers are operating. National funding initiatives and projects such as the Australian National Data Services’ Metadata Stores program have now provided both the infrastructure and the opportunity for development of capability within institutions. As eResearch and library professionals we have noted that just about every research group we deal with uses dropbox.com or similar file share and synchronisation services; this class of service is clearly the “killer app” for distributed teams working with filebased data. We have also observed that there is a gap in eResearch infrastructure between working data on fileshares and desktops, and the “proper” Research Data Repositories, eResearch tools, data capture tools and virtual laboratories now being established at universities.
Issues:
-
gap/ gulf between files and repositories
-
increasing need for researchers to cite data and the lack of easy routes to do this
-
The lack of decent low-end data packaging standards that are usable and accessible
-
The lack of plug-compatible, single-function solutions to build easy routes
-
Extreme range of data management use cases
-
The inability of big initiatives such as ANDS, NeCTAR, Cloustor+ to solve the micro use cases researchers face everyday
-
The lack of definitive and effective institutional name authority solutions for people, projects and data.
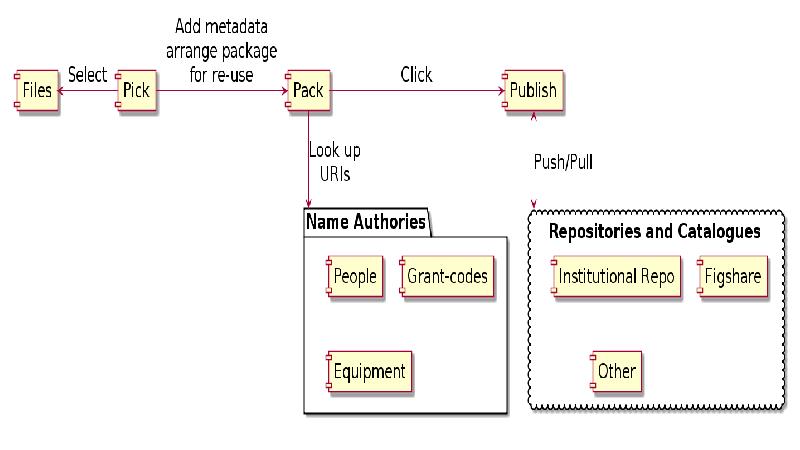
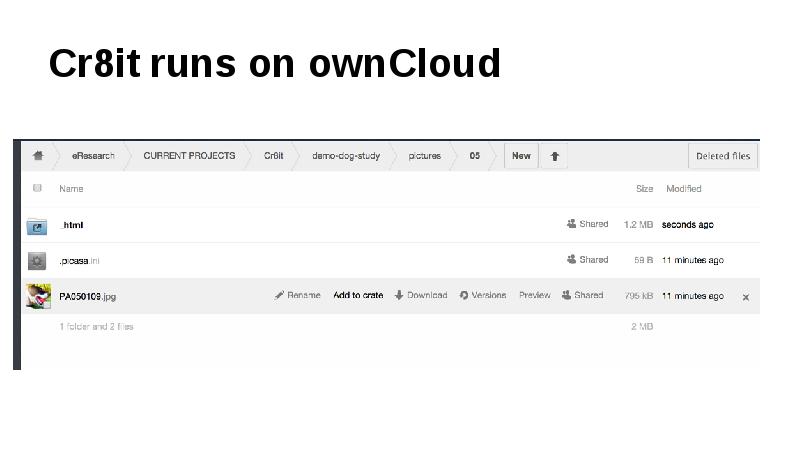
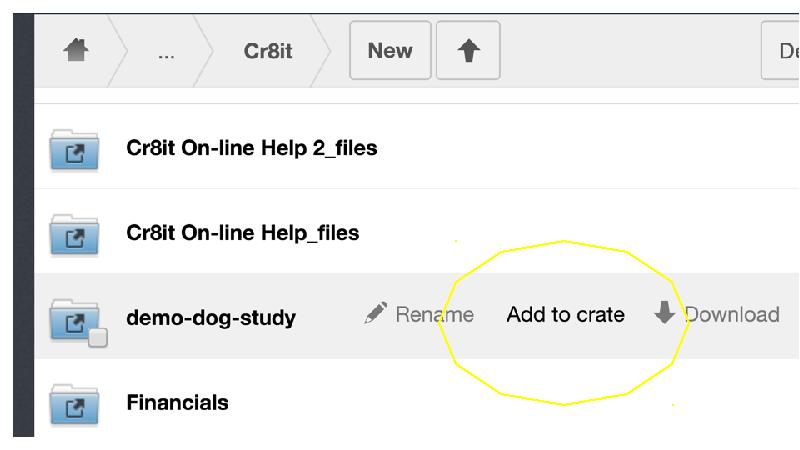
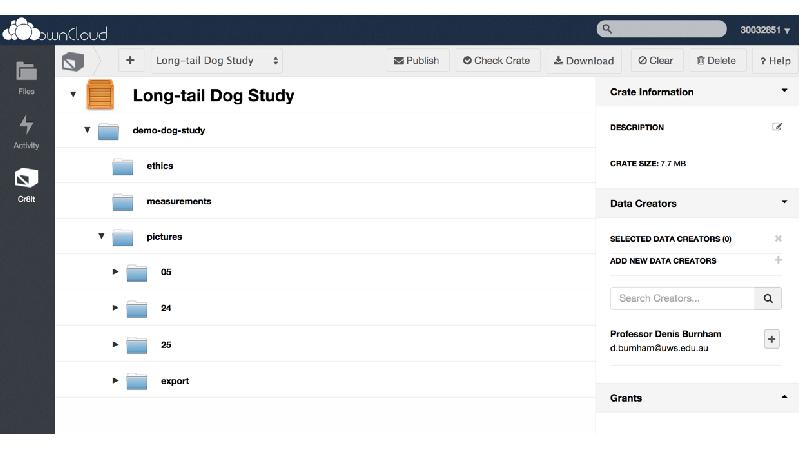
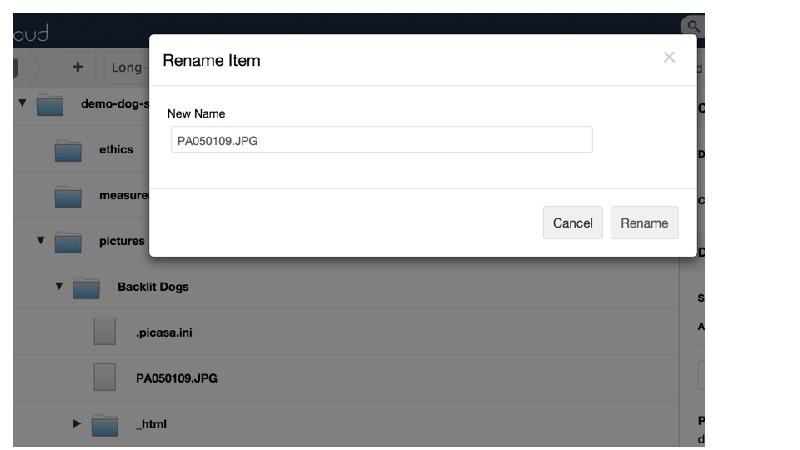
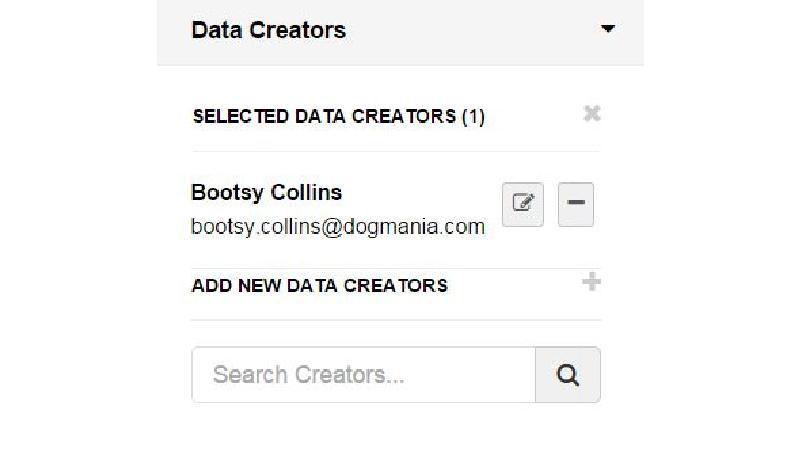
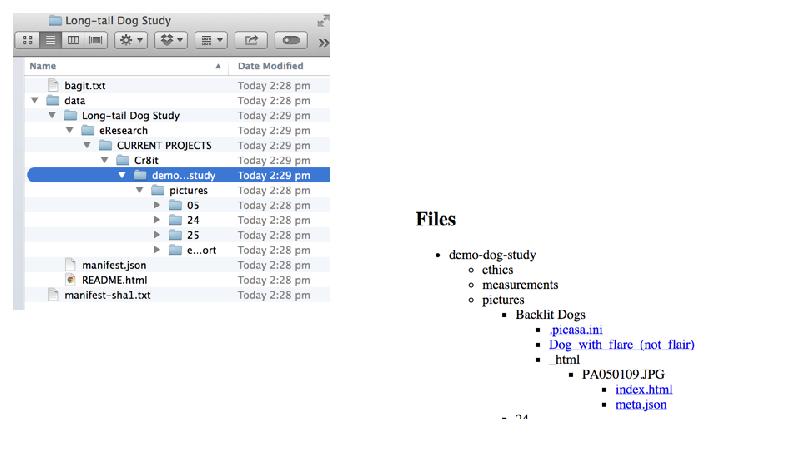
This gap is a huge barrier for researchers, as most tools require either laborious uploading of data, or forces researchers reorganise their data. Cr8it is designed to bridge that gap and eliminate the need to reorganise or move data, allowing it to be harvested, or ‘picked’ in situ. Our aim is to enable a seamless process, without disrupting the researcher’s workflow, to package research datasets, add metadata, and connect with data curation processes to publish a data description to the Research Data Commons and to appropriate discipline and other repositories. There are two main triggers for this: (a) publishing a research article, with supporting data and (b) archiving data at the end of a project.
Researchers now need to have end-to-end data management plans in place for all data, and all data must be archived appropriately to comply with the various codes and funding arrangements under which researchers are operating. National funding initiatives and projects such as the Australian National Data Services’ Metadata Stores program have now provided both the infrastructure and the opportunity for development of capability within institutions. As eResearch and library professionals we have noted that just about every research group we deal with uses dropbox.com or similar file share and synchronisation services; this class of service is clearly the “killer app” for distributed teams working with filebased data. We have also observed that there is a gap in eResearch infrastructure between workingdata on fileshares and desktops and the “proper” Research Data Repositories, eResearch tools, data capture tools and virtual laboratories now being established at universities.
This gap is a huge barrier for researchers, as most tools require either laborious uploading of data, or forces researchers reorganise their data. Cr8it is designed to bridge that gap and eliminate the need to reorganise or move data, allowing it to be harvested, or ‘picked’ in situ. Our aim is to enable a seamless process, without disrupting the researcher’s workflow, to package research datasets, add metadata, and connect with data curation processes to publish a data description to the Research Data Commons and to appropriate discipline and other repositories. There are two main triggers for this: (a) publishing a research article, with supporting data and (b) archiving data at the end of a project.